Ribosome, translation and cancer

Objectives
The Ribosome Translation and Cancer Team studies the emerging role of ribosomes and ribosome heterogeneity in cancer development. We identify cancer specific alterations of the ribosomes and of the ribosome production machinery with two aims: first to better understand the process of tumor development and second to apply this knowledge to the identification of novel biomarkers, to the development of novel therapeutic strategies to treat cancer, and to understanding response to chemotherapies. Our main pathologies of interest are breast, colon, lung and pancreatic cancers.
Projects
Our research comprises 6 themes, under the direction of Dr. J-J. Diaz:
- Therapeutic targeting of externalized proteins. (Dr. MA. Albaret)
- Nucleolin functions and targeting (Pr. P. Bouvet)
- Role of rRNA methylation in translation and therapeutic targeting (Dr. F. Catez)
- Alteration of ribosomes and translation by chemotherapies (Dr. N. Dalla Venezia)
- Ribosome and translational regulation of cell plasticity (embryonic stem cells, gliomas) (Dr. S. Durand)
- Role of rRNA methylation in cancer and clinical relevance (Dr. V. Marcel)
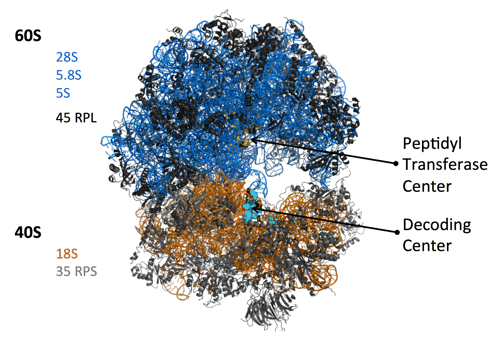
Ribosome biogenesis and cancer
Our group has a long-time interest on deregulations of ribosome biogenesis that are associated with pathological conditions, and in particular with tumor development and chemo-resistance. Ribosome biogenesis is a fundamental process of cell biology since it produces the ribosomes, the ribonucleoprotein complexes driving all protein synthesis of the cell. Recently, it has emerged that all ribosomes of a cell are not identical, and that the composition of a ribosome can impact on the translation activity, including quality of protein synthesis.
It is well known that ribosome biogenesis is over-activated in cancer cells, to cope with their hyper-proliferating state. As a consequence, morphology of nucleoli (the nuclear domains hosting ribosome biogenesis) is highly modified in cancer cells, and is one of the most distinctive characteristics of tumors grade. Genetic studies in various animal models brought evidence that modification of ribosome biogenesis, and consequently of ribosomes quality, could provide a survival benefit and thus participate in cancer development. Our group showed that in human cancers, ribosome production in qualitatively altered, and in particular that the methylation pattern of ribosomal RNAs (rRNA) is modified (Belin. S et al., PLoS ONE (2009); Marcel, V. et al., Cancer Cell (2013); Erales, J. et al. PNAS (2017) ; Marcel, V. et al., NAR Cancer (2020)).
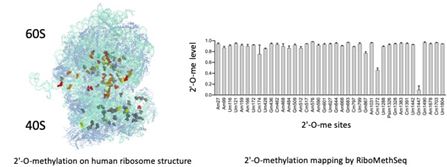
Role of 2′-O-methylation of ribosomal RNA in cancer
2′-O-methylation is the most abundant chemical modification of ribosomal RNA with 106 sites mapped in human ribosomes. Several studies including ours revealed that 2′-O-methylation contributes to the translational activity of the ribosome and impacts translation of mRNAs coding key cancer proteins (oncogenes, survival factors…). We are currently mapping 2′-O-methylation in various pathologies and cancer models to characterize 2′-O-methylation alteration and explore the potential of these alterationsin ribosome functions and as novel biomarkers. We use RiboMeth-Seq, a novel RNA-Seq based method which provides an exhaustive and quantitative mapping of 2′-O-methylation across all ribosomal RNAs (Erales, J. et al. PNAS (2017) ; Marcel, V. et al., NAR Cancer (2020)).
Our current projects include studying the role of variable 2′-O-Methylation sites on ribosome function, translation regulation and cellular phenotype.
Role of ribosome biogenesis master genes FBL and NCL in cancer biology
Cancer specific alterations of ribosome biogenesis are well established and contribute to both quantitative and qualitative alteration of the ribosome population in cancer cells. For instance, a wealth of data is available on RNA Pol I regulation by oncogenic and tumor suppressive pathways that coordinate ribosome biogenesis to the cell needs in healthy cells, and that abnormally increase ribosome biogenesis in cancer cells. In contrast, how downstream steps of ribosome maturation and assembly contribute to cancer specific ribosomes and to tumor phenotype has not been described yet. Fibrillarin (FBL) and Nucleolin (NCL) are two of the most important ribosome biogenesis factors and control both quantity and quality of the produced ribosome population. FBL is the 2′-O-methyltransferase of rRNAs, it is required for pre-rRNA cleavage into mature rRNAs and it stimulates RNA Pol I activity. NCL is required for RNA Pol I activity, pre-rRNA maturation and contributes to ribosome assembly. We explore the contribution and mechanistic roles of these two factors in several pathologies including breast cancer, leukemia and pancreatic cancers (Belin. S et al.,PLoS ONE (2009); Marcel, V. et al., Cancer Cell (2013), Marcel, V. et al.PLoS ONE (2017), Nguyen Van Long F, et al. Cancers, (2018), Kabirian-Dehkordi S, et al. Nanomedecine (2019) ; Nguyen Van Long F, et al. BMC Cancer 2022)).
Ribosomal heterogeneity and chemotherapy
Our team explores the contribution of ribosome variability to the resistance and mode of action of both non-targeted and targeted therapies, in colon and lung cancer. We recently focused on 5-Fluorouracil (5-FU), the most provided anti-cancer drug worldwide. 5-FU was firstly used for its anti-proliferative activity through deleterious effects on DNA. Only recently, attention was payed to incorporation of 5-FU metabolites into cellular RNAs. While 5-FU incorporation into RNA is up to 15,000 folds higher than into DNA, consequences on RNA function remain poorly understood. We showed that 5-FU induces a translational reprogramming however by a molecular mechanism that remains to be elucidated. We recently characterized the alteration of ribosome biogenesis and ribosome composition upon 5-FU treatment and we are exploring how these modifications contribute to treated cells translation regulation and phenotype (Bash-Imam, Z. et al. Oncotarget 2017 ; Therizols, G. et al. Nat. Commun. (2022) ; Chalabi-Dchar, M., NAR Cancer (2021)).
Our know-how
We develop and use a set of complementary technologies to describe at the molecular and cellular levels each step of ribosome biogenesis. Some are conventional biochemistry and cell biology methodologies (microscopy, reporter genes, tagged proteins…), and some are dedicated methodologies such as ribosome and nucleoli purification, polysome fractionation, or rRNA methylation analysis by RiboMethSeq. We collaborate with several teams with recognized expertise in Ribosome profiling, mass spectrometry and CryoEM analysis of ribosomes.
Openings
If you are interested to join the team, send your resume and motivation letter to Jean-Jacques Diaz (jean-jacques.diaz@lyon.unicancer.fr).
-
Jean-Jacques DIAZ
Team leader
jean-jacques.diaz@lyon.unicancer.fr
04 78 78 28 19Cheney A – 4th floor
Centre Léon Bérard