RibosOMICS Platform
Context
The RobosOMICS platform contribute to developments and provide access to high-throughput approaches dedicated to the analysis of rRNA translational regulation and epitranscriptomics.
Over the past 10 years, it has become clear that translation, which synthesizes proteins from mRNA via the ribosome, plays a key role in many physiological and pathological contexts, including cancer. This final stage in the regulation of gene expression contributes to the development of cancer phenotypes and progression, including resistance to anti-cancer therapies and metastatic dissemination.
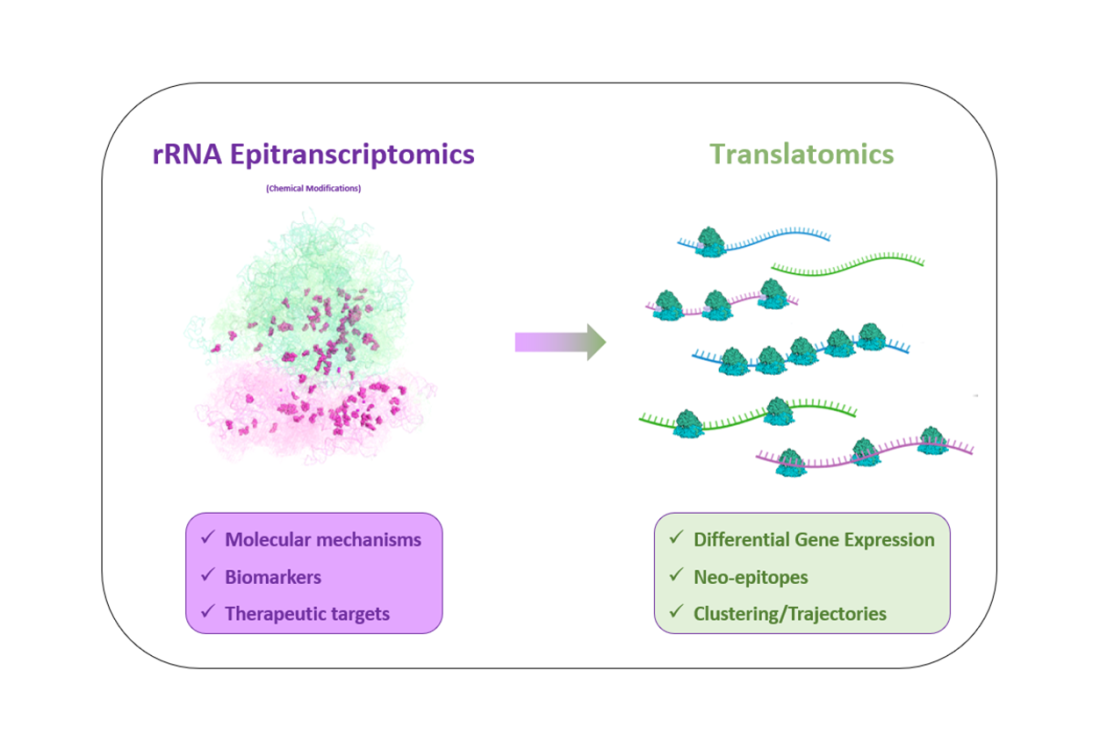
Today, understanding cancer through the prism of translational control highlights new molecular mechanisms in cancer, as well as new biomarkers and therapeutic targets, unsuspected until now. Among the key players in translation, the ribosome itself is emerging as a major regulator of translation. It turns out that ribosomes exhibit changes in their composition, notably in the chemical modifications of ribosomal RNAs (rRNAs), which affect their activity and selectivity towards certain mRNAs.
Operation
The RibosOMICS platform welcomes projects from CRCL/CLB and their collaborators, as well as external academics. For each project, it offers support in experimental design and handles technical implementation and bioinformatics analysis. The RibosOMICS platform covers the following approaches:
Epitranscriptomics of ribosomal RNAs : analysis of variations in the level of chemical modifications of rRNAs using biochemical approaches coupled with RNA-seqq :
- RiboMethSeq (data sheet): analysis of 2’O-ribose methylation of rRNAs
- Hydra(Ψ)seq (data sheet available soon) : rRNA pseudouridylation analysis
Translatomics :analysis of translation (differentially translated mRNAs and/or translational events) using biochemical approaches coupled with RNAseq or medium-throughput RTqPCR:
- PolySeq (data sheet available soon) : comparison of mRNAs associated with polysomes, i.e., ribosomes that are active and therefore undergoing translation. This approach enables the identification of differentially translated mRNAs.
- RiboSeq (data sheet available soon) : comparison of mRNAs protected by ribosomes. This approach allows the identification of differentially translated mRNAs, and under certain conditions, to identify translation defects and regulatory elements.
- RiboSTAMP (data sheet available soon) : comparison of mRNAs modified by the ribosome-associated enzyme APOBEC1. This approach enables the identification of differentially translated mRNAs.
Some approaches will be possible on a single-cell level :
- scRiboSeq (data sheet available soon)
- scRiboSTAMP (data sheet available soon)
To this end, and to simplify the flow of information, the RibosOMICS platform is responsible for managing the project and for administrative and experimental interactions with the other CRCL platforms. :
-
Operational manager :
Fleur BourdelaisScientific directors :
Virginie Marcel and Sébastien DurandTo apply to the RibosOMICS platform for a project, please refer to the downloadable procedure below and send it to us at the following e-mail address :
ribosomics@lyon.unicancer.fr